Today, give a try to Techtonique web app, a tool designed to help you make informed, data-driven decisions using Mathematics, Statistics, Machine Learning, and Data Visualization. Here is a tutorial with audio, video, code, and slides: https://moudiki2.gumroad.com/l/nrhgb. 100 API requests are now (and forever) offered to every user every month, no matter the pricing tier.
A few weeks ago in #112 and #120, I presented a few Python examples of Deep Quasi-Randomized ‘neural’ networks (QRNs). In this post, I will provide a detailed introduction to this new family of models, with examples in Python and R, and a preprint.
- Link to the preprint
- Link to a Jupyter Python notebook with a benchmark on 14 data sets: https://github.com/Techtonique/nnetsauce/blob/master/nnetsauce/demo/thierrymoudiki_20240519_deep_qrns.ipynb
At the basis of Deep QRNs are QRNs; nnetsauce’s CustomClassifier class objects, which, in turn, depend on a base Machine Learning model. This base learner could be any classifier, and in particular any scikit-learn classifier, or xgboost, or else. Here is how a CustomClassifier works, with the information flowing from left to right (forward pass only)
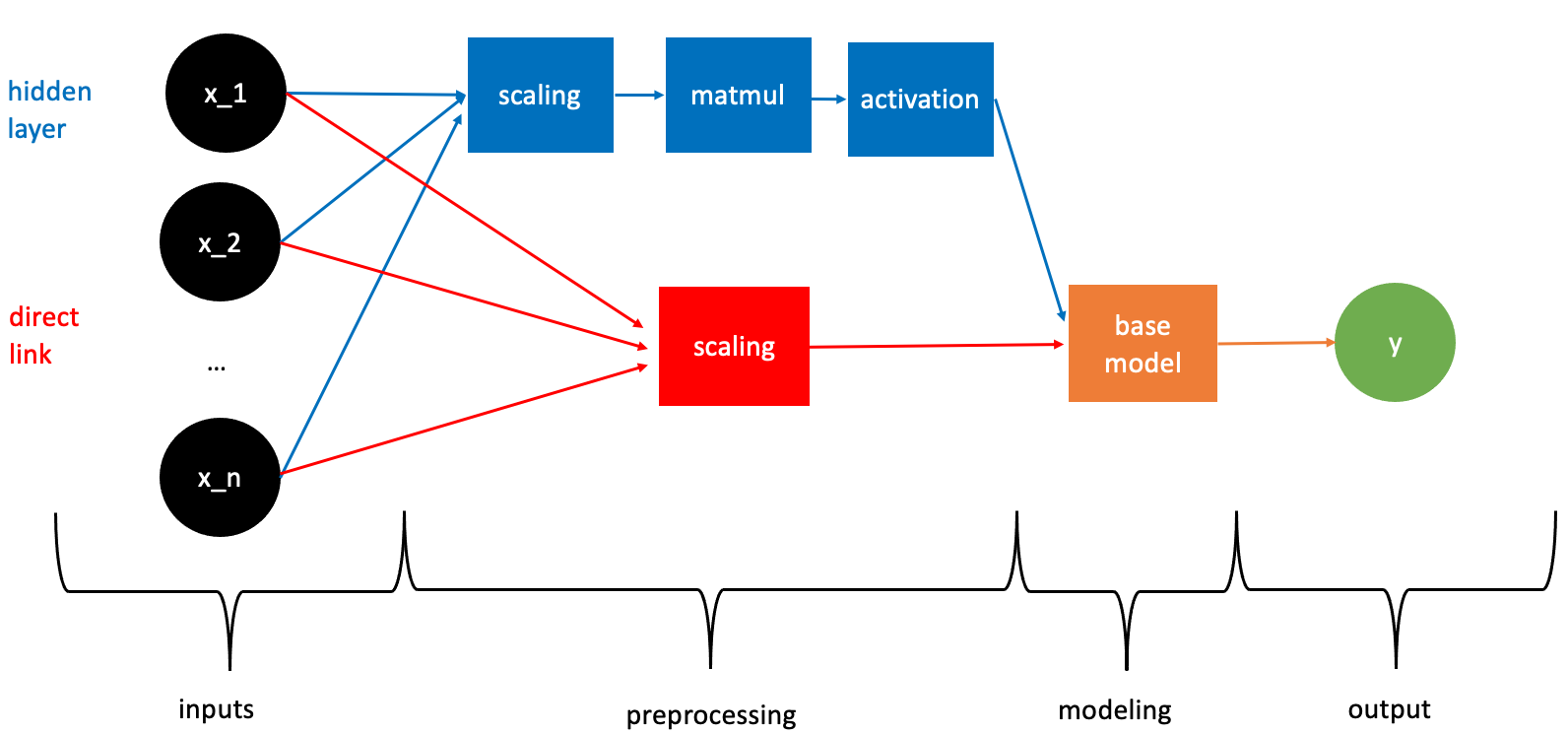
Deep QRNs arise from the QRN presented in this figure: in the case where we’d like to have a 3-layered deep QRN, the base Machine Learning model depicted in the figure can in turn be a QRN, and the obtained QRN can in turn be a QRN. Here are R examples:
Install nnetsauce
install.packages("nnetsauce", repos = c("https://techtonique.r-universe.dev", "https://cran.r-project.org"))
Load nnetsauce
library("nnetsauce")
iris data
library(datasets)
set.seed(123)
X <- as.matrix(iris[, 1:4])
y <- as.integer(iris$Species) - 1L
# split data into training and test sets
(index_train <- base::sample.int(n = nrow(X),
size = floor(0.8*nrow(X)),
replace = FALSE))
X_train <- X[index_train, ]
y_train <- y[index_train]
X_test <- X[-index_train, ]
y_test <- y[-index_train]
# base model is a Logistic Regression
obj2 <- sklearn$linear_model$LogisticRegressionCV()
# there are 3 layers in the deep model
obj <- DeepClassifier(obj2, n_layers = 3L)
# adjust the model
res <- obj$fit(X_train, y_train)
# accuracy, must be 1
print(mean(obj$predict(X_test)==y_test))
palmer penguins data
library(palmerpenguins)
data(penguins)
penguins_ <- as.data.frame(palmerpenguins::penguins)
replacement <- median(penguins$bill_length_mm, na.rm = TRUE)
penguins_$bill_length_mm[is.na(penguins$bill_length_mm)] <- replacement
replacement <- median(penguins$bill_depth_mm, na.rm = TRUE)
penguins_$bill_depth_mm[is.na(penguins$bill_depth_mm)] <- replacement
replacement <- median(penguins$flipper_length_mm, na.rm = TRUE)
penguins_$flipper_length_mm[is.na(penguins$flipper_length_mm)] <- replacement
replacement <- median(penguins$body_mass_g, na.rm = TRUE)
penguins_$body_mass_g[is.na(penguins$body_mass_g)] <- replacement
# replacing NA's by the most frequent occurence
penguins_$sex[is.na(penguins$sex)] <- "male" # most frequent
# one-hot encoding for covariates
penguins_mat <- model.matrix(species ~., data=penguins_)[,-1]
penguins_mat <- cbind.data.frame(penguins_$species, penguins_mat)
penguins_mat <- as.data.frame(penguins_mat)
colnames(penguins_mat)[1] <- "species"
y <- penguins_mat$species
X <- as.matrix(penguins_mat[,2:ncol(penguins_mat)])
n <- nrow(X)
p <- ncol(X)
set.seed(1234)
index_train <- sample(1:n, size=floor(0.8*n))
X_train <- X[index_train, ]
y_train <- factor(y[index_train])
X_test <- X[-index_train, ][1:5, ]
y_test <- factor(y[-index_train][1:5])
# base model is a Logistic Regression
obj2 <- nnetsauce::sklearn$linear_model$LogisticRegressionCV()
# there are 3 layers in the deep model
obj <- DeepClassifier(obj2, n_layers = 3L)
# adjust the model
res <- obj$fit(X_train, y_train)
# accuracy, must be 1
print(mean(obj$predict(X_test) == y_test))
It’s worth mentioning that the R version is a bit less stable than the Python version. Maybe because I’m not a reticulate superstar. I’m open to any suggestion/pull requests regarding this R port from the Python package.


Comments powered by Talkyard.